publications
2025
- PhD Thesis
 Symbols, Dynamics, and Maps: A Neurosymbolic Approach to Spatial CognitionNicole Sandra-Yaffa Dumont2025
Symbols, Dynamics, and Maps: A Neurosymbolic Approach to Spatial CognitionNicole Sandra-Yaffa Dumont2025
2024
- NICE
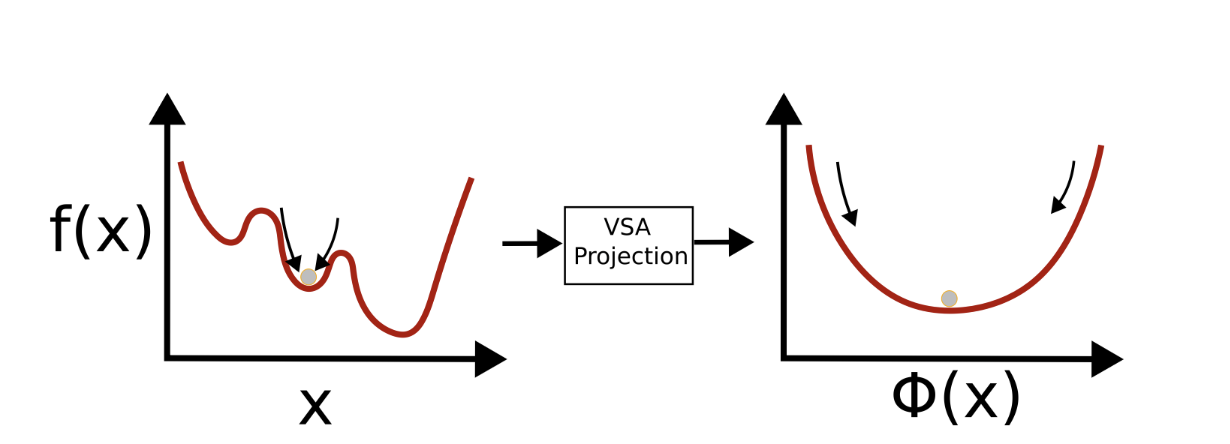 A Recurrent Dynamic Model for Efficient Bayesian OptimizationP Michael Furlong, Nicole Sandra-Yaffa Dumont, and Jeff OrchardIn 2024 Neuro Inspired Computational Elements Conference (NICE), 2024
A Recurrent Dynamic Model for Efficient Bayesian OptimizationP Michael Furlong, Nicole Sandra-Yaffa Dumont, and Jeff OrchardIn 2024 Neuro Inspired Computational Elements Conference (NICE), 2024Bayesian optimization is an important black-box optimization method used in active learning. An implementation of the algorithm using vector embeddings from Vector Symbolic Architectures was proposed as an efficient, neuromorphic ap- proach to solving these implementation problems. However, a clear path to neural implementation has not been explicated. In this paper, we explore an implementation of this algorithm expressed as recurrent dynamics that can be easily translated to neural populations, and present an implementation within the Lava programming framework for Intel’s neuromorphic computers. We compare the performance of the algorithm using different resolution representations of real-valued data, and demonstrate that the ability to find optima is preserved. This work provides a path forward to the implementation of Bayesian optimization on low-power neuromorphic computers, permitting the deployment of active learning techniques in low-power, edge computing applications.
@inproceedings{furlong2024recurrent, title = {A Recurrent Dynamic Model for Efficient Bayesian Optimization}, author = {Furlong, P Michael and Dumont, Nicole Sandra-Yaffa and Orchard, Jeff}, booktitle = {2024 Neuro Inspired Computational Elements Conference ({NICE})}, pages = {1--5}, year = {2024}, organization = {IEEE}, } - ICANN
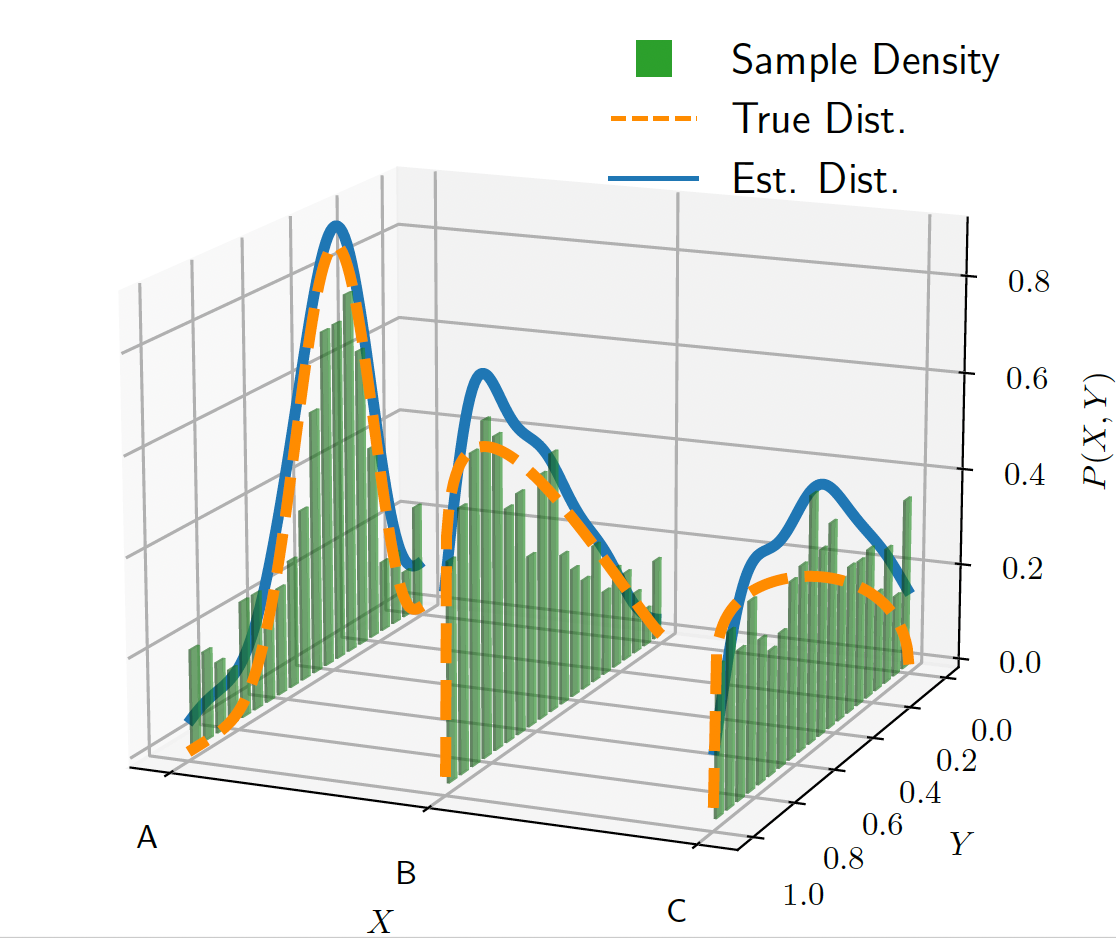 Biologically-Plausible Markov Chain Monte Carlo Sampling from Vector Symbolic Algebra-Encoded DistributionsP Michael Furlong, Kathryn Simone, Nicole Sandra-Yaffa Dumont, Madeleine Bartlett, Terrence C Stewart, Jeff Orchard, and Chris EliasmithIn International Conference on Artificial Neural Networks, 2024
Biologically-Plausible Markov Chain Monte Carlo Sampling from Vector Symbolic Algebra-Encoded DistributionsP Michael Furlong, Kathryn Simone, Nicole Sandra-Yaffa Dumont, Madeleine Bartlett, Terrence C Stewart, Jeff Orchard, and Chris EliasmithIn International Conference on Artificial Neural Networks, 2024Vector symbolic algebras (VSAs) are modelling frameworks that unify human cognition and neural network models, and some have recently been shown to be probabilistic models akin to Kernel Mean Em- beddings. Sampling from vector-embedded distributions is an important tool for turning distributions into decisions, in the context of cognitive modelling, or actions, in the context of reinforcement learning. However, current techniques for sampling from these distribution embeddings rely on knowledge of the kernel embedding or its gradient, knowledge which is problematic for neural systems to access. In this paper, we explore biologically-plausible Hamiltonian Monte Carlo Markov Chain sampling in the space of VSA encodings, without relying on any explicit knowledge of the encoding scheme. Specifically, we encode data using a Holographic Reduced Representation (HRR) VSA, sample from the encoded distributions using Langevin dynamics in the VSA vector space, and demonstrate competitive sampling performance in a spiking-neural network implementation. Surprisingly, while the Langevin dynamics are not constrained to the manifold defined by the HRR encoding, the generated samples contain sufficient information to reconstruct the target distribution, given an appropriate decoding scheme. We also demonstrate that the HRR algebra provides a straightforward conditioning operation. These results show that a generalized sampling model can explain how brains turn probabilistic latent representations into concrete actions in an encoding scheme-agnostic fashion. Moreover, sampling from vector embeddings of distributions permits the implementation of probabilistic algorithms, capturing uncertainty in cognitive models. We also note that the ease of conditioning distributions is particularly well-suited to reinforcement learning applications.
@inproceedings{furlong2024biologically, title = {Biologically-Plausible Markov Chain Monte Carlo Sampling from Vector Symbolic Algebra-Encoded Distributions}, author = {Furlong, P Michael and Simone, Kathryn and Dumont, Nicole Sandra-Yaffa and Bartlett, Madeleine and Stewart, Terrence C and Orchard, Jeff and Eliasmith, Chris}, booktitle = {International Conference on Artificial Neural Networks}, pages = {94--108}, year = {2024}, organization = {Springer}, }
2023
- ICCM
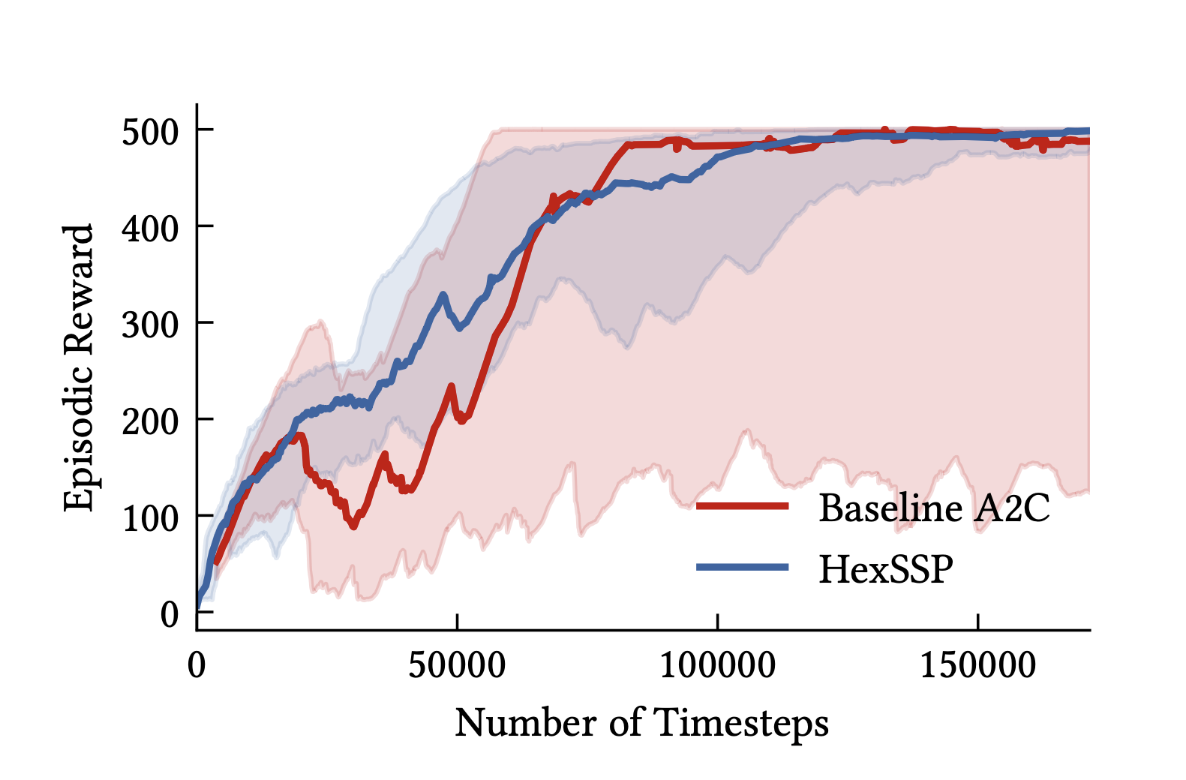 Improving reinforcement learning with biologically motivated continuous state representationsMadeleine Bartlett*, Kathryn Simone*, Nicole Sandra-Yaffa Dumont*, Chris Eliasmith, and Jeff OrchardJun 2023
Improving reinforcement learning with biologically motivated continuous state representationsMadeleine Bartlett*, Kathryn Simone*, Nicole Sandra-Yaffa Dumont*, Chris Eliasmith, and Jeff OrchardJun 2023Learning from experience, often formalized as Reinforcement Learning (RL), is a vital means for agents to develop successful behaviours in natural environments. However, while biological organisms are embedded in continuous spaces and continuous time, many artificial agents use RL algorithms that implicitly assume some form of discretization of the state space, which can lead to inefficient resource use and improper learning. In this paper we show that biologically motivated representations of continuous spaces form a valuable state representation for RL. We use models of grid and place cells in the Medial Entorhinal Cortex and hippocampus, respectively, to represent continuous states in a navigation task and in the CartPole control task. Specifically, we model the hexagonal grid structures found in the brain using Hexagonal Spatial Semantic Pointers, and combine this state representation with single-hidden-layer neural networks to learn action policies in an Actor-Critic framework. We demonstrate our approach provides significantly increased robustness to changes in environment parameters (travel velocity), and learns to stabilize the dynamics of the CartPole system with comparable mean performance to a deep neural network, while decreasing the terminal reward variance by more than 150x across trials. These findings at once point to the utility of leveraging biologically motivated representations for RL problems, and suggest a more general role for hexagonally-structured representations in cognition.
@conference{bartlett2023improving, title = {Improving reinforcement learning with biologically motivated continuous state representations}, author = {Bartlett, Madeleine and Simone, Kathryn and Dumont, Nicole Sandra-Yaffa and Eliasmith, Chris and Orchard, Jeff}, booktitle = {International Conference on Cognitive Modelling ({ICCM}) 2023}, year = {2023}, address = {Amsterdam, Netherlands}, month = jun, } - Frontiers in Neuro.
 Exploiting semantic information in a spiking neural SLAM systemNicole Sandra-Yaffa Dumont, P Michael Furlong, Jeff Orchard, and Chris EliasmithFrontiers in Neuroscience, Jun 2023
Exploiting semantic information in a spiking neural SLAM systemNicole Sandra-Yaffa Dumont, P Michael Furlong, Jeff Orchard, and Chris EliasmithFrontiers in Neuroscience, Jun 2023To navigate in new environments, an animal must be able to keep track of its position while simultaneously creating and updating an internal map of features in the environment, a problem formulated as simultaneous localization and mapping (SLAM) in the field of robotics. This requires integrating information from different domains, including self-motion cues, sensory, and semantic information. Several specialized neuron classes have been identified in the mammalian brain as being involved in solving SLAM. While biology has inspired a whole class of SLAM algorithms, the use of semantic information has not been explored in such work. We present a novel, biologically plausible SLAM model called SSP-SLAM—a spiking neural network designed using tools for large scale cognitive modeling. Our model uses a vector representation of continuous spatial maps, which can be encoded via spiking neural activity and bound with other features (continuous and discrete) to create compressed structures containing semantic information from multiple domains (e.g., spatial, temporal, visual, conceptual). We demonstrate that the dynamics of these representations can be implemented with a hybrid oscillatory-interference and continuous attractor network of head direction cells. The estimated self-position from this network is used to learn an associative memory between semantically encoded landmarks and their positions, i.e., an environment map, which is used for loop closure. Our experiments demonstrate that environment maps can be learned accurately and their use greatly improves self-position estimation. Furthermore, grid cells, place cells, and object vector cells are observed by this model. We also run our path integrator network on the NengoLoihi neuromorphic emulator to demonstrate feasibility for a full neuromorphic implementation for energy efficient SLAM.
@article{dumont2023exploiting, title = {Exploiting semantic information in a spiking neural SLAM system}, author = {Dumont, Nicole Sandra-Yaffa and Furlong, P Michael and Orchard, Jeff and Eliasmith, Chris}, journal = {Frontiers in Neuroscience}, volume = {17}, year = {2023}, publisher = {Frontiers Media SA}, doi = {10.3389/fnins.2023.1190515}, dimensions = {false}, } - Brain Sciences
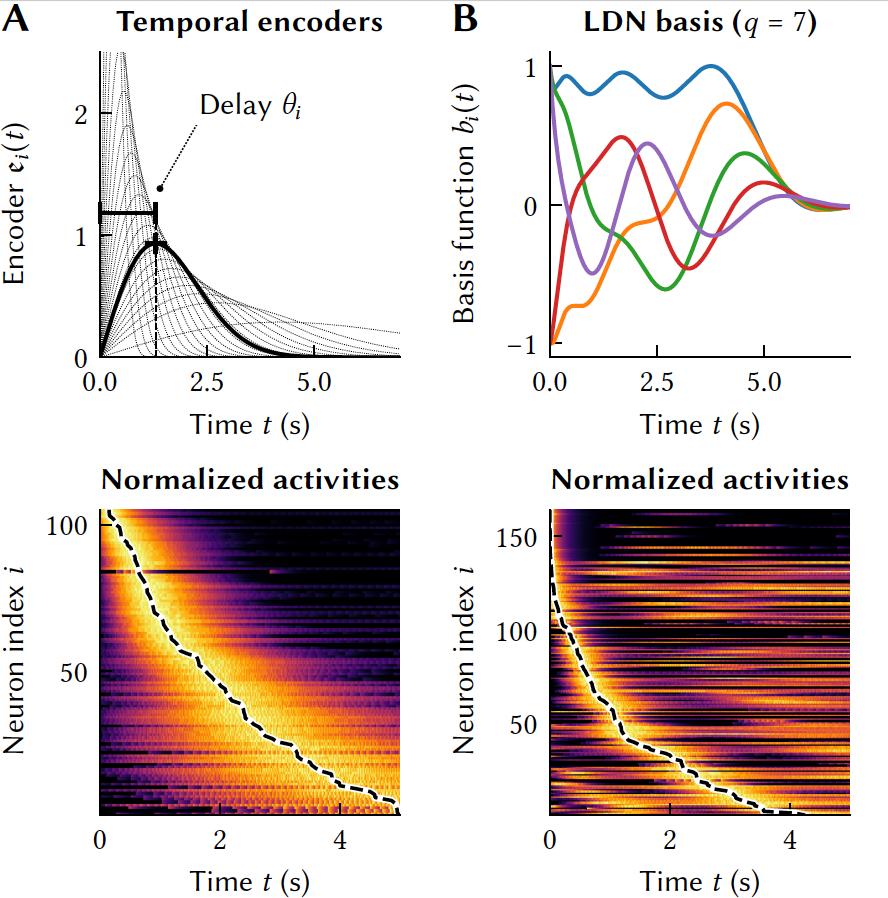 Biologically-Based Computation: How Neural Details and Dynamics Are Suited for Implementing a Variety of AlgorithmsNicole Sandra-Yaffa Dumont, Andreas Stöckel, P Michael Furlong, Madeleine Bartlett, Chris Eliasmith, and Terrence C StewartBrain Sciences, Jun 2023
Biologically-Based Computation: How Neural Details and Dynamics Are Suited for Implementing a Variety of AlgorithmsNicole Sandra-Yaffa Dumont, Andreas Stöckel, P Michael Furlong, Madeleine Bartlett, Chris Eliasmith, and Terrence C StewartBrain Sciences, Jun 2023The Neural Engineering Framework (Eliasmith & Anderson, 2003) is a long-standing method for implementing high-level algorithms constrained by low-level neurobiological details. In recent years, this method has been expanded to incorporate more biological details and applied to new tasks. This paper brings together these ongoing research strands, presenting them in a common framework. We expand on the NEF’s core principles of (a) specifying the desired tuning curves of neurons in different parts of the model, (b) defining the computational relationships between the values represented by the neurons in different parts of the model, and (c) finding the synaptic connection weights that will cause those computations and tuning curves. In particular, we show how to extend this to include complex spatiotemporal tuning curves, and then apply this approach to produce functional computational models of grid cells, time cells, path integration, sparse representations, probabilistic representations, and symbolic representations in the brain.
@article{dumont2023biologically, title = {Biologically-Based Computation: How Neural Details and Dynamics Are Suited for Implementing a Variety of Algorithms}, author = {Dumont, Nicole Sandra-Yaffa and St{\"o}ckel, Andreas and Furlong, P Michael and Bartlett, Madeleine and Eliasmith, Chris and Stewart, Terrence C}, journal = {Brain Sciences}, volume = {13}, number = {2}, pages = {245}, year = {2023}, publisher = {MDPI}, doi = {10.3390/brainsci13020245}, dimensions = {false}, }
2022
- ICCM
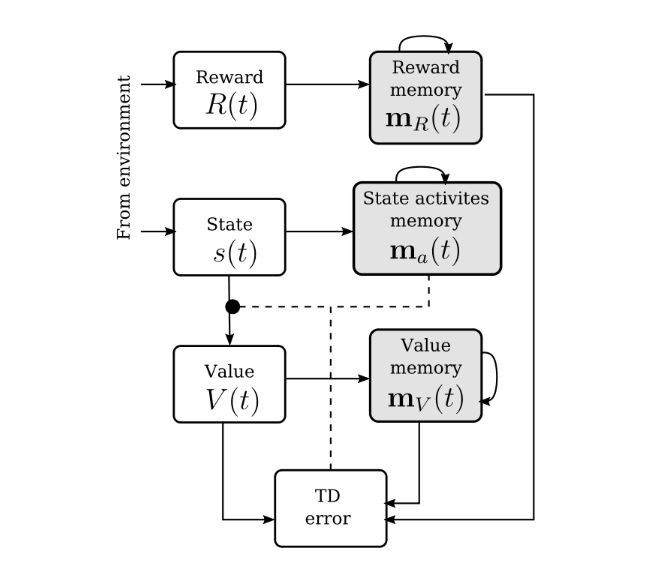 Biologically-Plausible Memory for Continuous Time Reinforcement LearningMadeleine Bartlett*, Nicole Sandra-Yaffa Dumont*, Michael P. Furlong, and Terry C. StewartJul 2022
Biologically-Plausible Memory for Continuous Time Reinforcement LearningMadeleine Bartlett*, Nicole Sandra-Yaffa Dumont*, Michael P. Furlong, and Terry C. StewartJul 2022Reinforcement learning, and particularly Temporal Difference learning, has been inspired by, and offers insights into, the mechanisms underlying animal learning. An ongoing challenge to providing biologically realistic models of learning is the need for algorithms that operate in continuous time and can be implemented with spiking neural networks. This paper presents a novel approach to Temporal Difference learning in continuous time – TD(θ). This approach relies on the use of Legendre Delay Networks for storing information about the past that will be used to update the value function. A comparison of the discrete-time TD(n) and continuous TD(θ) rules on a simple spatial navigation RL task in a largely non-spiking network is presented, and the theoretical implications and avenues for future work are discussed.
@conference{bartlett2022, author = {Bartlett, Madeleine and Dumont, Nicole Sandra-Yaffa and Furlong, Michael P. and Stewart, Terry C.}, title = {Biologically-Plausible Memory for Continuous Time Reinforcement Learning}, booktitle = {International Conference on Cognitive Modelling ({ICCM}) 2022}, year = {2022}, address = {Toronto, Canada}, month = jul, } - CogSci
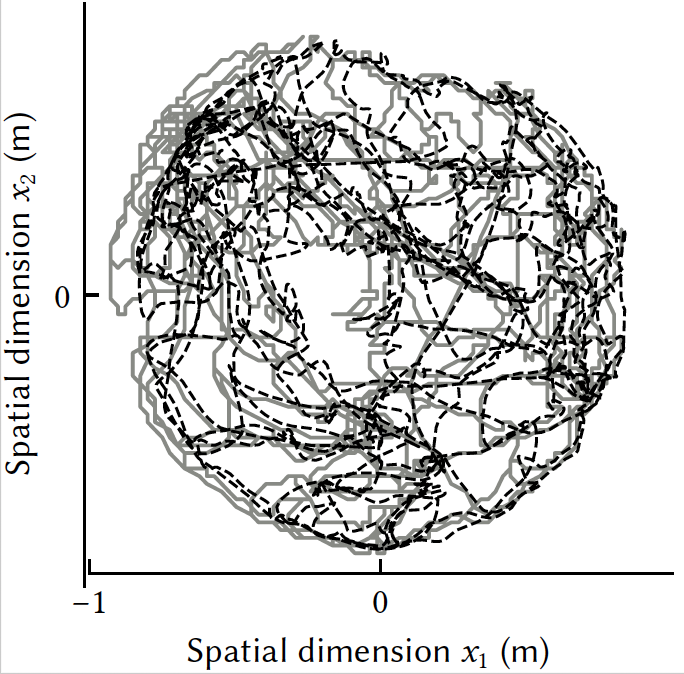 A model of path integration that connects neural and symbolic representationNicole Sandra-Yaffa Dumont, Jeff Orchard, and Chris EliasmithIn Proceedings of the Annual Meeting of the Cognitive Science Society, Jul 2022
A model of path integration that connects neural and symbolic representationNicole Sandra-Yaffa Dumont, Jeff Orchard, and Chris EliasmithIn Proceedings of the Annual Meeting of the Cognitive Science Society, Jul 2022Path integration, the ability to maintain an estimate of one’s location by continuously integrating self-motion cues, is a vital component of the brain’s navigation system. We present a spiking neural network model of path integration derived from a starting assumption that the brain represents continuous variables, such as spatial coordinates, using Spatial Semantic Pointers (SSPs). SSPs are a representation for encoding continuous variables as high-dimensional vectors, and can also be used to create structured, hierarchical representations for neural cognitive modelling. Path integration can be performed by a recurrently-connected neural network using SSP representations. Unlike past work, we show that our model can be used to continuously update variables of any dimensionality. We demonstrate that symbol-like object representations can be bound to continuous SSP representations. Specifically, we incorporate a simple model of working memory to remember environment maps with such symbol-like representations situated in 2D space.
@inproceedings{dumont2022, title = {A model of path integration that connects neural and symbolic representation}, author = {Dumont, Nicole Sandra-Yaffa and Orchard, Jeff and Eliasmith, Chris}, booktitle = {Proceedings of the Annual Meeting of the Cognitive Science Society}, volume = {44}, number = {44}, year = {2022}, publisher = {Cognitive Science Society}, address = {Toronto, ON}, }
2021
- Comp. Econ.
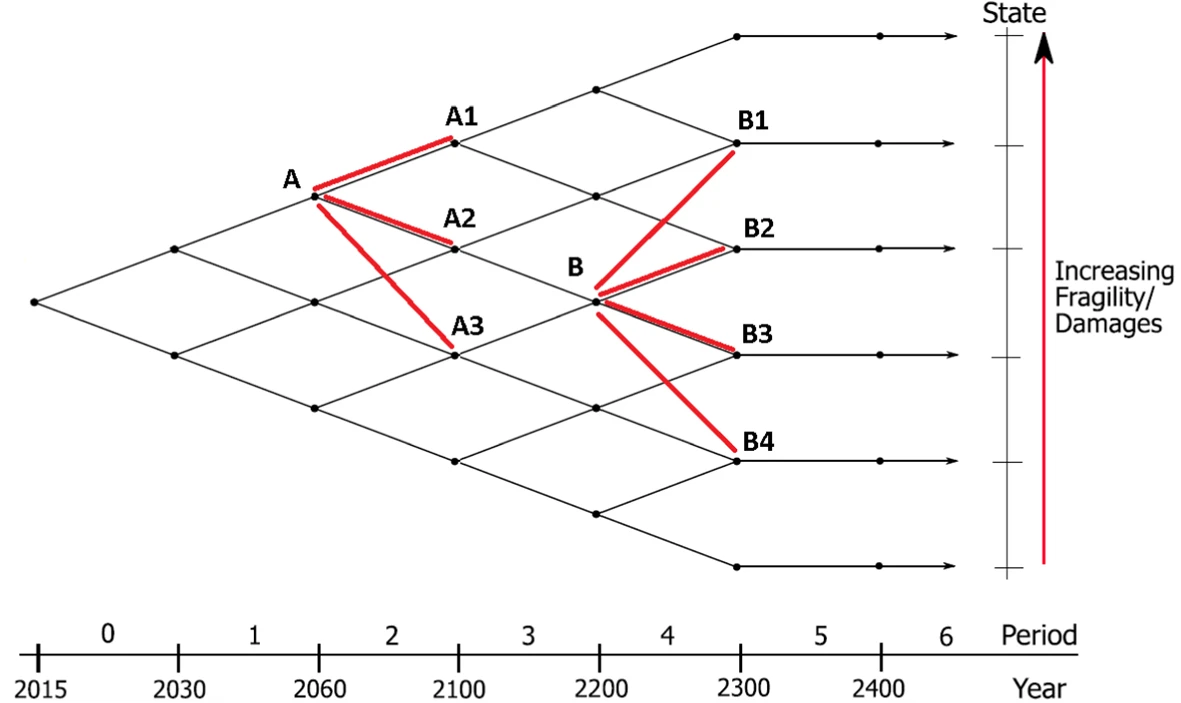 Optimal Pricing of Climate RiskThomas F Coleman, Nicole Sandra-Yaffa Dumont, Wanqi Li, Wenbin Liu, and Alexey RubtsovComputational Economics, Jul 2021
Optimal Pricing of Climate RiskThomas F Coleman, Nicole Sandra-Yaffa Dumont, Wanqi Li, Wenbin Liu, and Alexey RubtsovComputational Economics, Jul 2021@article{coleman2021optimal, title = {Optimal Pricing of Climate Risk}, author = {Coleman, Thomas F and Dumont, Nicole Sandra-Yaffa and Li, Wanqi and Liu, Wenbin and Rubtsov, Alexey}, journal = {Computational Economics}, pages = {1--34}, year = {2021}, publisher = {Springer}, doi = {10.1007/s10614-021-10179-6}, } - Neural Computation
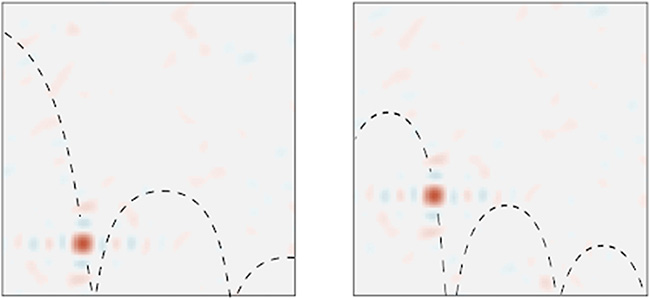 Simulating and Predicting Dynamical Systems With Spatial Semantic PointersAaron R Voelker, Peter Blouw, Xuan Choo, Nicole Sandra-Yaffa Dumont, Terrence C Stewart, and Chris EliasmithNeural Computation, Jul 2021
Simulating and Predicting Dynamical Systems With Spatial Semantic PointersAaron R Voelker, Peter Blouw, Xuan Choo, Nicole Sandra-Yaffa Dumont, Terrence C Stewart, and Chris EliasmithNeural Computation, Jul 2021While neural networks are highly effective at learning task-relevant representations from data, they typically do not learn representations with the kind of symbolic structure that is hypothesized to support high-level cognitive processes, nor do they naturally model such structures within problem domains that are continuous in space and time. To fill these gaps, this work exploits a method for defining vector representations that bind discrete (symbol-like) entities to points in continuous topological spaces in order to simulate and predict the behavior of a range of dynamical systems. These vector representations are spatial semantic pointers (SSPs), and we demonstrate that they can (1) be used to model dynamical systems involving multiple objects represented in a symbol-like manner and (2) be integrated with deep neural networks to predict the future of physical trajectories. These results help unify what have traditionally appeared to be disparate approaches in machine learning.
@article{voelker2021simulating, title = {Simulating and Predicting Dynamical Systems With Spatial Semantic Pointers}, author = {Voelker, Aaron R and Blouw, Peter and Choo, Xuan and Dumont, Nicole Sandra-Yaffa and Stewart, Terrence C and Eliasmith, Chris}, journal = {Neural Computation}, volume = {33}, number = {8}, pages = {2033--2067}, year = {2021}, publisher = {MIT Press One Rogers Street, Cambridge, MA}, doi = {10.1162/neco_a_01410}, dimensions = {false}, } - COSYNE
 Spiking neural network model of simultaneous localization and mapping with Spatial Semantic PointersNicole Sandra-Yaffa Dumont, Terry C. Stewart, and Chris EliasmithFeb 2021
Spiking neural network model of simultaneous localization and mapping with Spatial Semantic PointersNicole Sandra-Yaffa Dumont, Terry C. Stewart, and Chris EliasmithFeb 2021@conference{dumont2021, author = {Dumont, Nicole Sandra-Yaffa and Stewart, Terry C. and Eliasmith, Chris}, title = {Spiking neural network model of simultaneous localization and mapping with Spatial Semantic Pointers}, booktitle = {Computational and Systems Neuroscience ({COSYNE}) 2021}, year = {2021}, address = {Online}, month = feb, }
2020
- CogSci
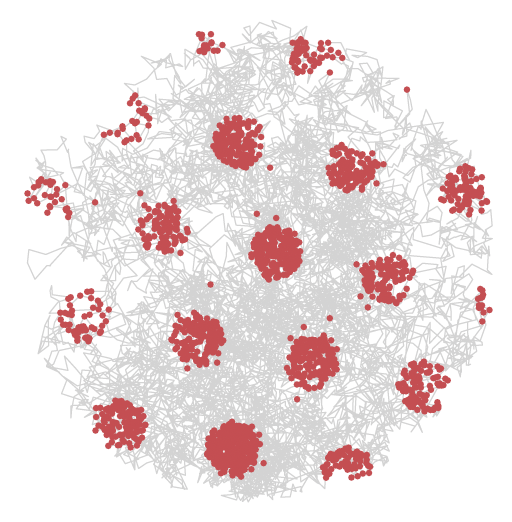 Accurate representation for spatial cognition using grid cellsNicole Sandra-Yaffa Dumont, and Chris EliasmithIn 42nd Annual Meeting of the Cognitive Science Society, Feb 2020
Accurate representation for spatial cognition using grid cellsNicole Sandra-Yaffa Dumont, and Chris EliasmithIn 42nd Annual Meeting of the Cognitive Science Society, Feb 2020Spatial cognition relies on an internal map-like representation of space provided by hippocampal place cells, which in turn are thought to rely on grid cells as a basis. Spatial Semantic Pointers (SSP) have been introduced as a way to represent continuous spaces and positions via the activity of a spiking neural network. In this work, we further develop SSP representation to replicate the firing patterns of grid cells. This adds biological realism to the SSP representation and links biological findings with a larger theoretical framework for representing concepts. Furthermore, replicating grid cell activity with SSPs results in greater accuracy when constructing place cells. Improved accuracy is a result of grid cells forming the optimal basis for decoding positions and place cell output. Our results have implications for modelling spatial cognition and more general cognitive representations over continuous variables.
@inproceedings{dumont2020, title = {Accurate representation for spatial cognition using grid cells}, author = {Dumont, Nicole Sandra-Yaffa and Eliasmith, Chris}, booktitle = {42nd Annual Meeting of the Cognitive Science Society}, year = {2020}, pages = {2367--2373}, publisher = {Cognitive Science Society}, address = {Toronto, Canada}, }